A meta-data standard
A meta-data standard
 for mesh based data ...
for mesh based data ...
 and particle data sets.
and particle data sets.
 Clean Common Structure
Clean Common Structure
 ... and Domain Specific Extensions
... and Domain Specific Extensions
How to avoid confusion between actual low-level data sets & attributes (HDF5 speak), variables & attributes (ADIOS speak), files & groups/folders and the actual physical quantities?
openPMD naming convention- discretized vector field \(\vec F(\vec r)\) on a mesh
- discretized scalar field \(T(\vec r)\) on a mesh
- particle property position \(\vec r_i\)
- ...
- unitSI: conversion factor to common unit system
- unitDimension: parsable dimensionality
- time / timeUnitSI: iteration != time
- automate description of units
- powers of the 7 (SI) base measures
- e.g., `V/m` is length1.0 mass1.0 time-3.0 electrical current-1.0 thermodynamic temperature0.0 ammount of substance0.0 luminous intensity0.0: `[1.0, 1.0, -3.0, -1.0, 0.0, 0.0, 0.0]`
- (if your record can be scaled in a general normalized way, choose a reference and write a note in the `comment` attribute)
- geometry (+ parameters)
- grid: spacing, global offset, axis labels
- data order (C/Fortran)
- position (on grid)
- position
- position offset (can be constant)
Optional:
- id
- / ... / meshes / E /
- x
- y
- z
- / ... / meshes /
- \(T\)
- / ... / particles / electrons / position /
- x
- y
- z
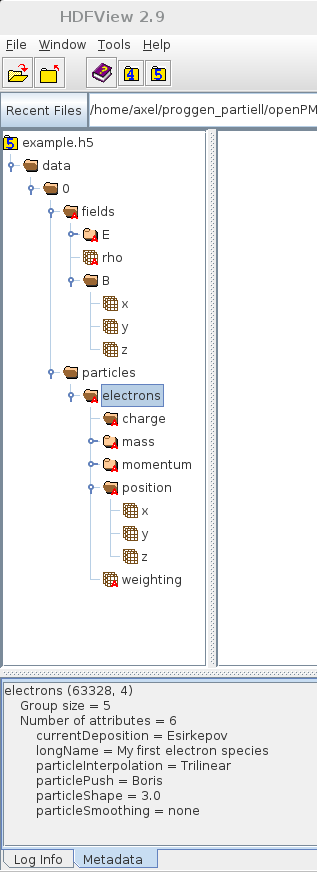
- / ... / particles / electrons
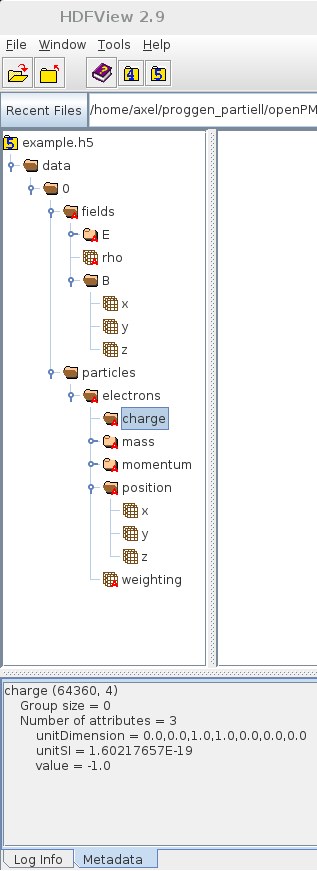
- charge
- / ... / particles / electrons
- charge ← might be very large
- unitSI
- charge ← might be very large
- / ... / particles / electrons
- charge
- value ← few bytes
- shape
- unitSI
- charge
- / ... / particles / electrons
- position
- x + unitSI
- y + unitSI
- z
- value, shape
- unitSI
- position
- parallel, community file formats: writing/reading based on MPI & MPI-I/O
- examples:
- PHDF5 .h5 (parallel/strided, uncompressed)
- ADIOS .bp (aggregated, compressed)
 Particle Patches: Honor Decomposition
Particle Patches: Honor Decomposition
 Particle Patches: Disjoint Particle Sets
Particle Patches: Disjoint Particle Sets
 Particle Patches: [Offset:Offset+Count]
Particle Patches: [Offset:Offset+Count]
 Particle Patches: (Spatial) Hyperrectangles
Particle Patches: (Spatial) Hyperrectangles
In principle and everywhere*: a human-readable comment (text) attribute is encouraged for everything not covered by the standard.
* reserved for each group and data set
Comment AttributeopenPMD defines a minimal set of attributes.
You can always add more attributes and records!
openPMD is a not exclusiveopenPMD defines a minimal set of attributes, e.g.
- openPMD: identifier
- basePath: prefix, currently fixed to `/data/`
- meshesPath: relative sub-group, e.g., `meshes/`
- particlesPath: relative sub-group, e.g., `particles/`
- author: My Name <email@example.com>
- software: e.g., PIConGPU
- softwareVersion: e.g., 0.1.0
- date: 2015-12-02 17:48:42 +0100
tools/ $ ./createExamples_h5.py$ ./checkOpenPMD_h5.py -i example.h5 --EDPIC
Warning: Attribute softwareVersion (recommended) does NOT exist in `/`!
Found 1 iteration(s)
Iteration 0 : found 2 meshes
Iteration 0 : found 1 particle species
Result: 0 Errors and 1 Warning.- meta-data parsing
- file-format independent (ADIOS & HDF5)
- object oriented: meshes, particles & iterations
- Python API: openPMD object aware
- GUI: IPython Notebook (interactive, remote)
- ideal for investigating 1-2D data (or slices)
- modular: e.g., domain specific analysis chains
- VisIt, Paraview (GUI, Python)
- libSplash (C/C++)
- numpy-like parallel access
- read and write support
- based on zeroMQ / Jupyter notebook
- openPMD-viewer numpy-like parallel access
- read and write support
- GUI: Jupyter notebook widgets
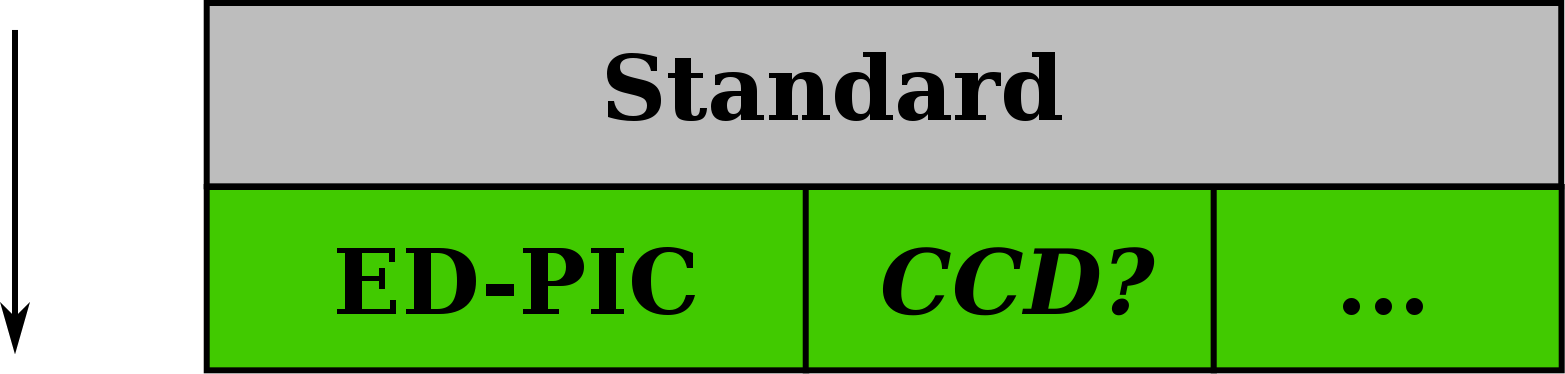 Extensions: Domain-Specific Additions
Extensions: Domain-Specific Additions
- electro-dynamic and electro-static PIC
- additional attributes
- naming conventions for records
- Experimental data: CCD images, interferograms, ...
- Simulations: MD, FEM, ...
- record patches and AMR support
- irregular cartesian grids
- more mesh types: if required